In the features that I’ve coded so far, I’m deliberately telling the cell to do specific things or creating rules. But Cells should learn all this upon evolving, not during their own lifetime. So, slowly, each of the traits of the cell needs to be governed by genes and then set to be randomly activated upon mutation. But I’ll need to first code all these traits, and then leave them to a chance mutation.
For different genes, turning on and off a mutation should be sticky. So if a mutation has 1% chance of being turned on, then for each new generation that has that mutation, it has a 1% chance of being turned off. That way, if the mutation is beneficial, then there will be more and more members having that mutation. If it is not beneficial, then fewer and fewer cells will be born with that mutation
Cells are learning to move because if you can move, you get access to food and so you can live longer, and produce more children (and pass on your genes to them)
Watched the youtube video I programmed some creatures. They Evolved. - YouTube and the simulator is very similar to what I’m creating. I can use some of the ideas from there and incorporate into this one. The most compelling one is coding a simple neural network for the brain. I’ve never coded a neural network by hand and creating one from scratch is compelling. One of my concerns when programming the simulator was that I was getting tempted to code certain behaviours in the cells. For example, I had written the logic for the cell to try to move when the food has depleted. But if one of the sensory inputs is ‘quantify of food’ and one of the actions the call can take is move then the neural network can ‘learn’ to move when the food has depleted. With the neural network, I should only program the interface from the cell to ask information about its environment, and for it to be able to take actions in the environment. In between these, all the logic should be learnt by the cell’s brain.
But I do have some issues with I programmed some creatures. They Evolved. - YouTube . The main one is that at the end of each generation, organisms are distributed spatially suddenly. This isn’t what would happen in real life. If we’re doing a simulation like this, then nothing else should be dependent on the ‘location’ of a gene. For example, it should be able to kill anyone on the grid irrespective of the proximity. What I’d like to do is do a continuous simulation we don’t stop and distribute the cells spatially.
Discovered major flaw
I’ve found a major flaw in my simulator: my data structures are the same as the ‘Game of Life’ implementation and essentially I’m traversing a matrix and updating each cell. But if an organism is allowed to move, then I can’t really do this! Say I’m traversing the the matrix left to right (inner loop), and top to bottom(outer loop) and a cell decides to move right. Then I’ll be computing that cell again in the iteration of the inner loop. This means, the same cell can be computed multiple times in the same iteration. For cells that can move from one block to another, and definitely for multi-cellular organisms, I’ll need to traverse the list of cells and not the list of blocks on the grid.
Fixed base simulator and resolved several bugs with the old implementation. The new graphs look much more stable

Thanks to my google searches, youtube has been recommending me videos of evolutionary simulations non-stop. Watched Simulating the Evolution of a Zombie Pandemic (Virus simulation) - YouTube and loved it. Really good way to show the importance of mutations in evolution in a human setting that’s easy to understand. I have been thinking of simulating parasites in my simulator and may want to do something similar.
Another good video is Interactive Evolution Simulator - YouTube . I didn’t know that the creator actually built an interactive simulator for the blobs on Primer - YouTube .
I reached out to the creator of I programmed some creatures. They Evolved. - YouTube asking him more about his simulator and he mentioned the following links which are quite interesting:
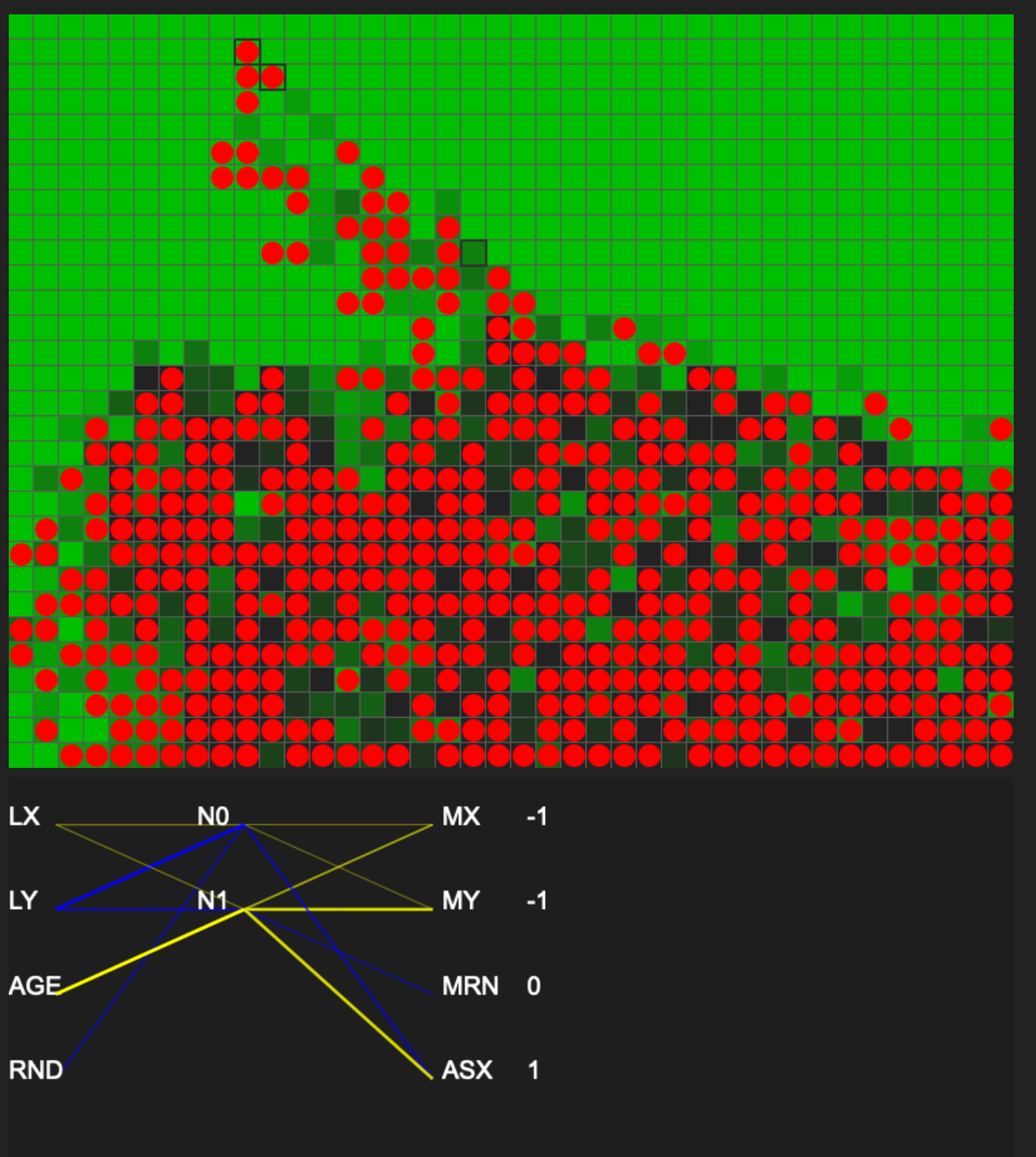
Started coding a typical neural network based on vector multiplication of matrices, but switched the implementation to use the encoding as described in I programmed some creatures. They Evolved. - YouTube. The way it has been done in this video is less optimal, but it is closer to how evolution works in real life. I would prefer for my implementation to be close to real-life evolution but my simulation is going to run in a browser with limited resources. So I am going back to the vector-multiplication method of implementing and computing ANNs and might come to the gene-encoding method later. The vector multiplication method is almost the same as Evolution Simulator with Neural Networks in Unity! - YouTube where for mutations, I will randomly select different weights or biases for the connections. I don’t have biases implemented at the moment and starting with just weights.
Cannot rescrict number of genes
One disadavantge of using the vec-mul ANN Vs the gene-encoding ANN is that with the former, it won’t be easily possible to restrict the number of genes. I could enforce it by the following logic:
Movement
When there are multiple output nodes in the neural network that are realted to movement, then how should the movement actually happen? Say the ‘move_random’ output is fired at the same time as ‘move_x’. One way is to start with the highest probability output and keep traversing the outputs. But I could also convert the outputs into 0s and 1s, that way I can use the pre-calculated result to decide where the cell should finally move.
I’m now plotting the neural network for each selected cell. And the behavior of each cell is predictable after looking at its network. But with the NN visualization, I’m more confident that there aren’t any major logic bugs in the simulator at the moment. Next up:
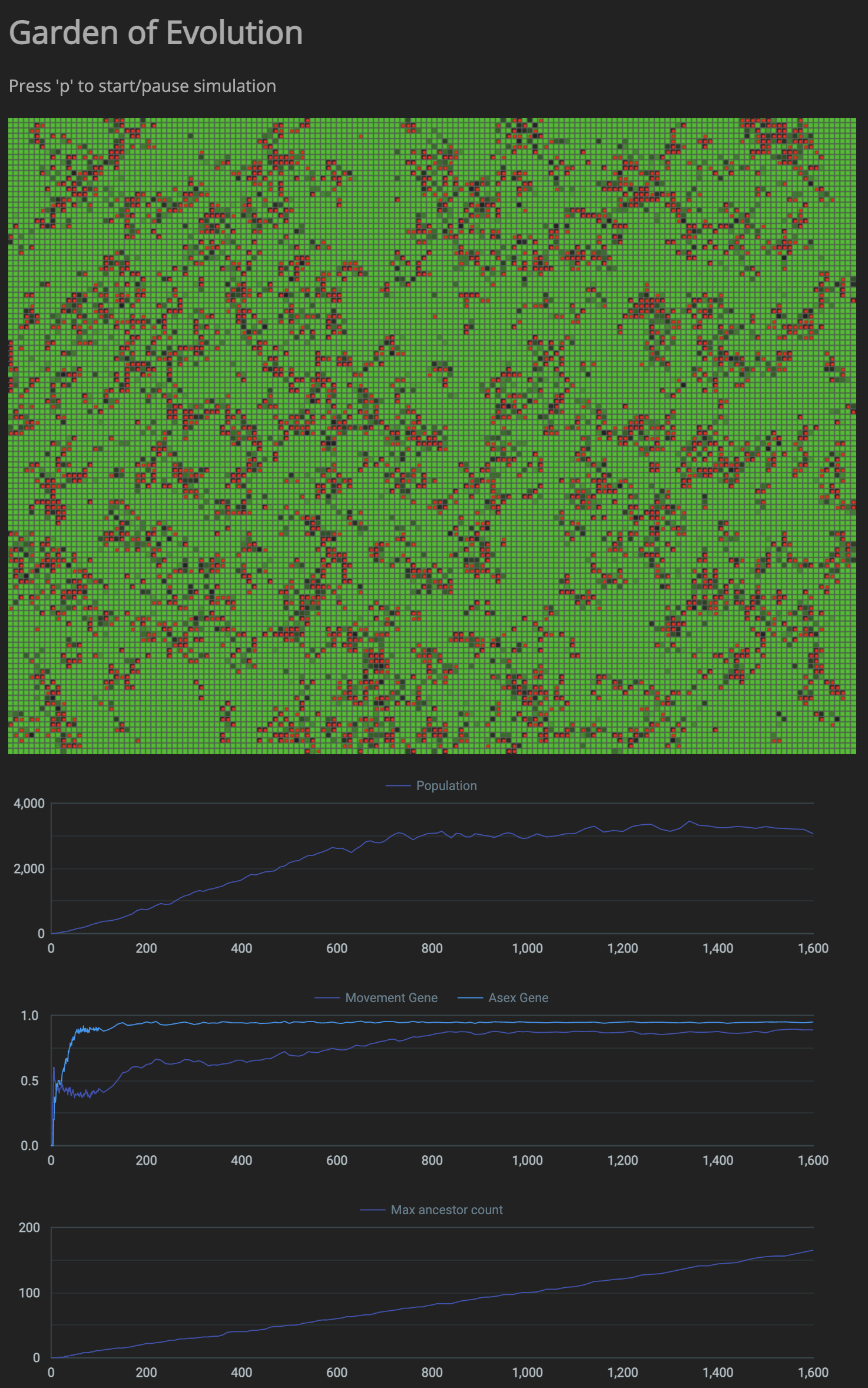
As expected, the simulation is just showing random results without the possibility of mutation. I’ve implemented mutation and now the only way to find out the effect of mutation is to see if certain mutations become dominant. So I need to plot the graph of the number of species that exist at any time, and in those species find the relative occurance of individual species. Working on plotting those graphs.
Displaying dominant species count too.
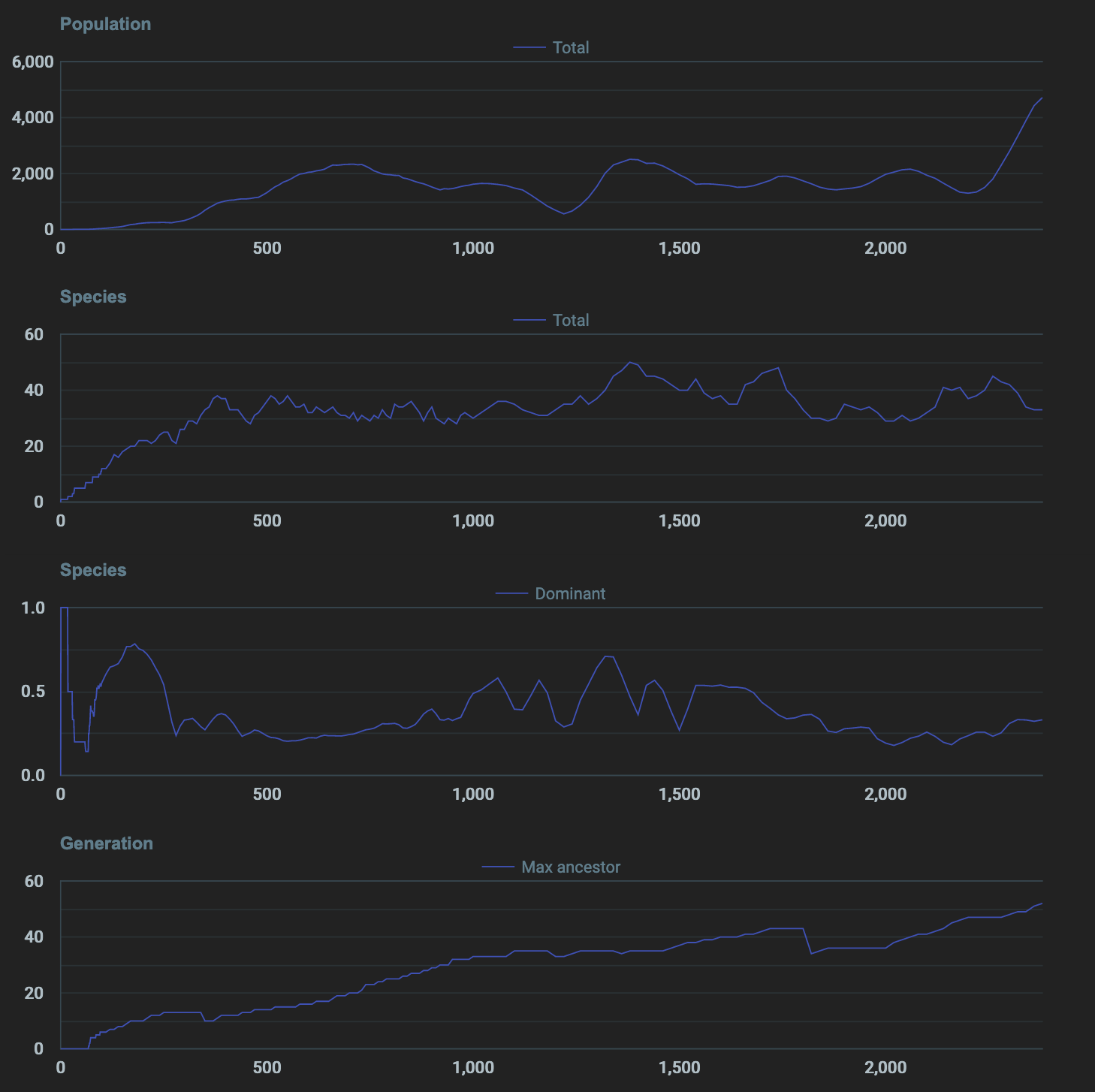
Not seeing enough mutations
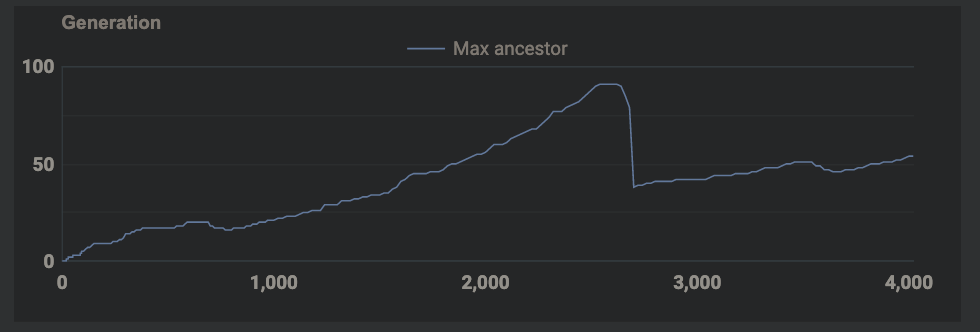
In the current environment, the max ancestor count is increasing very slowly and then plateauing after that. This means that the cells aren’t really able to mutate over several hundreds of generations. In order to see genetic learning to be successful there needs to be an environment that fosters reproduction as much as possible. Things I could do to reach several hundreds of generations in a continuous environment are:
Implemented both these features.
Problems with resetting the simulation
On the UI front, I also need a reliable to reset the environment. When resizing the canvas something very strange is happening and it feels like the relative distances on the HTML canvas are getting messed up causing the particles to speed up or slow down.
Fixed by prompting a reload
Tried the fixes above, max ancestor count is still not increasing. Genes aren’t getting a chance to last hundreds of generations. I need to figure out how to provide a way for them to naturally survive many generations instead of being artificially moved to next generation and made to reproduce (like in other simulation examples I’ve seen so far)
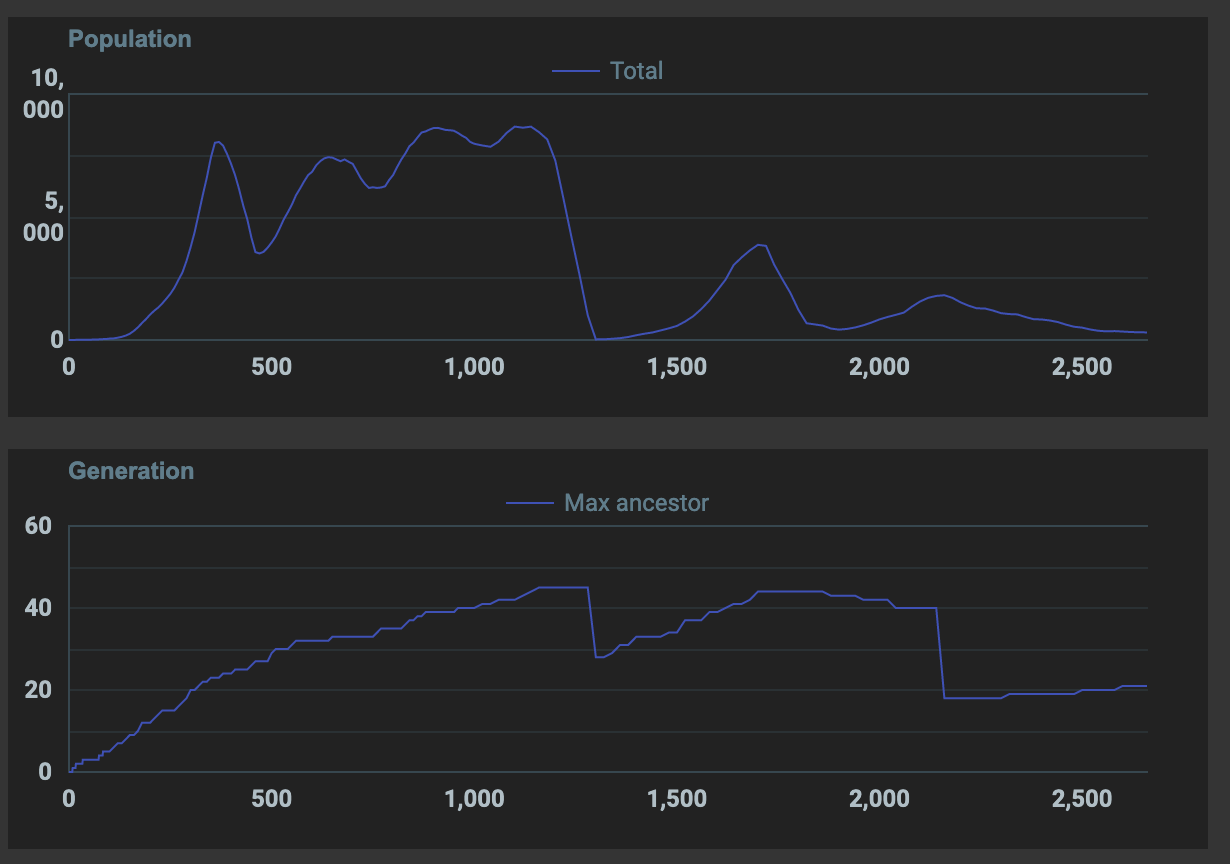
Thinking about this more, what’s preventing genes from surviving?
How can I prevent overcrowding?
How can I promote reproduction and mutation?
I’ll need to play with these parameters
Update I played with parameters but I’m just not able to cause a specific dominant species to evolve and cause the genes to survive enough number of generations. Thoughts for later: